As of September 30th, 2019:
9.29 billion docking jobs have been submitted.
As of August 12th, 2019:
9.11 billion docking jobs have been submitted.
As of April 23rd, 2019:
8.57 billion docking jobs have been submitted.
As of March 20th, 2019:
7.78 billion docking jobs have been submitted.
As of February 15th, 2019:
7.51 billion docking jobs have been submitted.
As of January 18th, 2019:
7.24 billion docking jobs have been submitted.
As of November 29th, 2018:
6.79 billion docking jobs have been submitted.
Dockings against the capsid proteins (binding pockets 1 and 2) were the last jobs submitted.
Thank you all for your help!
As of October 4th, 2018:
5.91 billion docking jobs have been submitted
As of July 31st, 2018:
In total, we have submitted almost 5 billion docking jobs, which involved 427 different target sites. Our initial screens used an older library of 6 million commercially available compounds, and our current experiments utilize a newer library of 30.2 million compounds. We have already received approximately 4.6 billion of these results on our server (there is some lag time between when the calculations are performed on your volunteered machines and when we get the results, since all of the results per “package” of approximately 10,000 – 50,000 different docking jobs need to be returned to World Community Grid, re-organized, and then compressed before sending them to our server).
Thus far, the > 80,000 volunteers who have donated their spare computing power to OpenZika have given us > 51,471 CPU years worth of docking calculations, at a current average of 64.5 CPU years per day!
Thank you all very much for your help!
As of April 10th, 2018:
5.04 billion docking jobs have been submitted
As of Nov 10th, 2017:
4.4 billion docking jobs have been submitted
As of May 17th, 2017:
3.5 billion docking jobs have been submitted
2 billion results have been received
As of February 1st, 2017:
2.56 billion docking jobs have been submitted
Virtual screening of 427 different target sites versus 6 million compounds
As of November 18th, 2016:
We have received 100% of the results for Experiments 1A, 1B (NS5 class), 2A, 2B, and 2C (NS3 helicase class).
We have received 46% of the results for Experiment 3A (NS1 class).
1A = 132 million docking jobs
1B = 42 million
2A = 354 million
2B = 360 million
2C = 54 million
3A = we have received 160 million out of 348 million.
In total, we have submitted ~ 1.6 billion docking jobs, and we have already received ~ 1.1 billion results.
As of June 19th, 2016:
The results from the first 3 experiments involve 84 different target sites (74 unique targets, mostly from Dengue Virus, HCV, Yellow Fever Virus, Japanese Encephalitis Virus and Zika Virus). Thus, as of June 19th, 2016, we have received 254 million different docking results (in approximately 2 months).
Experiment 1A = Virtual Screening against the NS5 class (first binding site)
The non-structural protein 5 (NS5) of flaviviruses is the most conserved amongst the viral proteins. The N-terminal domain of NS5 possesses the methyltransferase and guanylyltransferase activities necessary for forming mature RNA cap structures.
Thus, as of July 19th, 2016, we have received 99.75% of the results for Experiment 1A, corresponding to 125,688,000 docking jobs.
| Batch Range | LibraryNickname |
| 1 – 21 | Drugs_and_Leads_FromPrestwick_ |
| 22 – 4956 | Enamine_Full_Library |
| 4957 – 8127 | Vitas-M Labs_Full_Library |
| 8128 – 10269 | ChemBridge_Full_Library |
| 10270 – 11340 | Asinex_Full_Library |
| 11341 – 12012 | NCI_Full_Library |
| 12013 – 19026 | Otava_Full_Library (note: only 1,000 ligands/batch) |
Batches for all other libraries have ~10,000 ligands/batch
Targets employed to virtual screening:
| NS5 Hepatitis C virus complexed with Sofosbuvir and RNA |
| NS5 Homology Model of Zika virus minimized |
| RNA polymerase Hepatitis C virus, Chain A |
| RNA polymerase, Hepatitis C virus, Chain B |
| RNA polymerase, Hepatitis C virus,complexed with RNA and Mn, Chain A |
| RNA polymerase, Hepatitis C virus, complexed with RNA and Mn, Chain B |
| RNA polymerase, West Nile virus |
| NS5B Hepatitis C virus complex with 1,5 benzodiazepine inhibitor 6, Chain A, Configuration B |
| NS5B Hepatitis C virus complex with 1,5 benzodiazepine inhibitor 6, Chain A |
| NS5B Hepatitis C virus complex with 1,5 benzodiazepine inhibitor 6, Chain B |
| NS5B Hepatitis C virus complex with 1,5 benzodiazepine inhibitor 6, Chain C |
| RNA polymerase, Hepatitis C virus, without RNA |
| RNA polymerase,Hepatitis C virus, with RNA |
| NS5 Japanese encephalitis virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE, Chain A |
| NS5 Japanese encephalitis virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE, Chain B |
| NS5 Dengue virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE |
| NS5 Dengue virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE |
| NS5 Dengue virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE and GTP |
| NS5 Hepatitis C virus complexed with Sofosbuvir and without RNA |
| NS5 Homology Model of Zika virus align to template 4k6m |
| NS5 Homology Model of Zika virus align to template 4v0r |
These images below show a “positive control” experiment. This experiment demonstrates that we can accurately predict/reproduce the specific way that a known inhibitor binds to this important drug target, NS5 class.

Positive control for NS5 class of target: Alpha test
crystallographic binding mode: ball-and-stick, dark purple carbon atoms;
predicted binding mode: green carbons atoms


Positive control for NS5 class of target: Beta test
Positive control for NS5 class of target: Beta test
crystallographic binding mode: purple carbon atoms,
predicted binding mode: cyan carbon atoms
Experiment 1B = Virtual Screening against the NS5 class (refined NS5 and second binding site)
The protein structure NS5, obtained by homology modeling, was refined using the server KobaMIN. This refinement provided us a better structure, decreasing the number steric overlaps, bad bonds/angles, Ramachandran outliers, twisted peptides…
The second binding site of NS5 is related to the ligand S-Adenosyl Methionine (SAM) position in the binding site.
Thus, as of July 19th, 2016, we have received 87% of the results for Experiment 1B, corresponding to ~ 42 million docking jobs.
| Batch Range | Library Nickname |
| 19027 – 19034 | Drugs_and_Leads_FromPrestwick_MicroSourceSpectrum_FDA_and_NIHclinicalCollection |
| 19035 – 20914 | Enamine_Full_Library |
| 20915 – 22122 | Vitas-M Labs_Full_Library |
| 22123 – 22938 | ChemBridge_Full_Library |
| 22939 – 23346 | Asinex_Full_Library |
| 23347 – 23602 | NCI_Full_Library |
| 23603 – 23874 | Otava_Full_Library (note: now has 10,000 ligands/batch) |
Targets employed to virtual screening:
| NS5 Model Refined |
| NS5 Japanese encephalitis virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE, Chain A, binding site 2 |
| NS5 Japanese encephalitis virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE, Chain B, binding site 2 |
| NS5 Dengue virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE, binding site 2 |
| NS5 Dengue virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE, binding site 2 |
| NS5 Dengue virus, complexed with S-ADENOSYL-L-HOMOCYSTEINE and GTP, binding site 2 |
| NS5 Homology Model of Zika virus align to template 4k6m, binding site 2 |
| NS5 Homology Model of Zika virus align to template 4v0r, binding site 2 |
Experiment 2A = Virtual Screening against the NS3 helicase class (first binding site).
The NS3 helicase domain exhibits intrinsic nucleoside triphosphatase activity (upon stimulation by RNA), which then provides the chemical energy to unwind viral RNA replication intermediates to facilitate replication of the viral genome together with RNA-dependent RNA polymerase (NS5). Given its essential role in genome replication, ZIKV NS3 helicase could be an attractive target for drug development against ZIKV.
The NS3 helicase crystallographic structure was solved in May 12th 2016 (PDB ID: 5JMT)
The NS3 helicase structure revealed a conserved triphosphate pocket critical for nonspecific hydrolysis of nucleoside triphosphates across multiple flavivirus species. A positive-charged tunnel has been identified in the viral helicase, which is potentially responsible for accommodating the RNA.
The first binding site of NS3 is related to the nucleic acid binding site.
Thus, as of July 19th, 2016, we have received 26% of the results for Experiment 2A, corresponding to ~ 86 million docking jobs.
| Batch Range | LibraryNickname |
| 23875 – 23929 | Drugs_and_Leads_FromPrestwick_MicroSourceSpectrum_FDA_and_NIHclinicalCollection |
| 23930 – 36854 | Enamine_Full_Library |
| 36855 – 45159 | Vitas-M Labs_Full_Library |
| 45160 – 50769 | ChemBridge_Full_Library |
| 50770 – 53574 | Asinex_Full_Library |
| 53575 – 55334 | NCI_Full_Library |
| 55335 – 57204 | Otava_Full_Library (note: now has 10,000 ligands/batch) |
Targets employed to virtual screening:
| 5jmt ZIKV NS3helicase A |
| 5jmt ZIKV NS3helicase B |
| 1yks YFV NS3hlcs |
| 2jlu DV4 NS3hlcs ssRNA chA |
| 2jlu DV4 NS3hlcs ssRNA chB |
| 2jlv_DV4_NS3hlcs_ssRNA_chA_A |
| 2jlv_DV4_NS3hlcs_ssRNA_chA_B |
| 2jlv_DV4_NS3_ssRNAdelMn_chA_A |
| 2jlv_DV4_NS3_ssRNAdelMn_chA_B |
| 2jlv_DV4_NS3_ssRNAdelMn_chB_A |
| 2jlv_DV4_NS3_ssRNAdelMn_chB_B |
| 2jlw_Deng4_NS3_ssRNA2_chA |
| 2jlw_Deng4_NS3_ssRNA2_chB |
| 2jlx_DV4_NS3hlcs_RNAdelMnChA |
| 2jlx_DV4_NS3hlcs_RNAdelMnChB |
| 2jlx_DV4_NS3hlcs_ssRNA_chA |
| 2jlx_DV4_NS3hlcs_ssRNA_chB |
| 2jly_DV4_NS3hlcs_ssRNA_chA_A |
| 2jly_DV4_NS3hlcs_ssRNA_chA_B |
| 2jly_DV4_NS3hlcs_ssRNAdelMn_chA_A |
| 2jly_DV4_NS3hlcs_ssRNAdelMn_chA_B |
| 2jly_DV4_NS3hlcs_ssRNAdelMn_chB |
| 2jlz_DV4_NS3hlcs_ssRNA_chA |
| 2jlz_DV4_NS3_ssRNAdelMn_chA |
| 2whx_DV_NS3hlcsDelMn_PRwasBnd |
| 2whx_DV_NS3hlcs_PRwasPepBnd |
| 2z83_JEV_NS3hlcs |
| 1a1v_HCV_NS3hlcs_ssDNA |
| 3kqh_HCV_NS3hlcs_ssDNA_chA |
| 3kqh_HCV_NS3hlcs_ssDNA_chB |
| 3kqk_HCV_NS3hlcs_chA |
| 3kqk_HCV_NS3hlcs_chB |
| 3kql_HCV_NS3hlcs_chnA |
| 3kql_HCV_NS3hlcs_chnB |
| 3kql_HCV_NS3hlcsDelMg_chA |
| 3kql_HCV_NS3hlcsDelMgChB |
| 3kqn_HCV_NS3hlcs_ssDNA_DelMn |
| 3kqn_HCV_NS3hlcs_ssDNA |
| 3o8b_HCV_NS3hlcs_PR_chA |
| 3o8b_HCV_NS3hlcs_PR_chB |
| 3o8c_HCV_NS3_Hlcs_PR_chB |
| 3o8c_HCV_NS3_ssRNA_wPR_chA |
| 4wxp_HCV_NS3helcs_A |
| 1nkt_Mtb_SecA_chnA |
| 1nkt_Mtb_SecA_chnB |
| 1nkt_Mtb_SecA_DelMg_chnA |
| 1nkt_Mtb_SecA_DelMg_chnB |
| 4uaq_Mtb_SecA2 |
| 3mla_Anthrax_BaNadDApochB |
| 3mla_Anthrax_BaNadD_chA_A |
| 3mla_Anthrax_BaNadD_chA_B |
| 4es6_PAeru_HemD_A |
| 4es6_PAeru_HemD_B |
| 4nl4_Klebp_PriA_A |
| 4nl4_Klebp_PriA_B |
Results available for the scientific community
Results of Virtual Screening of ZIKV NS3 helicase (PDB ID: 5jmt) with the databases: Prestwick + MicroSource Spectrum + FDA approved + NIH clinical collection set
ZIKA_000023875_x5jmt_ZIKV_NS3helicase_A_results.tgz
The ZIKV NS3 helicase structure is available below.
Experiment 2B = Virtual Screening against the NS3 helicase class (second binding site)
The second binding site of NS3 is related to the ATP binding site.
| Batch Range | LibraryNickname |
| 57205 – 57259 | Drugs_and_Leads_FromPrestwick_MicroSourceSpectrum_FDA_and_NIHclinicalCollection |
| 57260 – 70184 | Enamine_Full_Library |
| 70185 – 78489 | Vitas-M Labs_Full_Library |
| 78490 – 84099 | ChemBridge_Full_Library |
| 84100 – 86904 | Asinex_Full_Library |
| 86905 – 88664 | NCI_Full_Library |
| 88665 – 90534 | Otava_Full_Library (note: now has 10,000 ligands/batch) |
Targets employed to virtual screening:
| 5jmt_ZIKV_NS3helicase_A_s2 |
| 5jmt_ZIKV_NS3helicase_B_s2 |
| 2jlu_DV4_NS3hlcs_ssRNA_chA_s2 |
| 2jlu_DV4_NS3hlcs_ssRNA_chB_s2 |
| 2jlv_DV4_NS3hlcs_ssRNA_chA_A_s2 |
| 2jlv_DV4_NS3hlcs_ssRNA_chA_B_s2 |
| 2jlv_DV4_NS3_ssRNAdelMn_chA_A_s2 |
| 2jlv_DV4_NS3_ssRNAdelMn_chA_B_s2 |
| 2jlv_DV4_NS3_ssRNAdelMn_chB_A_s2 |
| 2jlv_DV4_NS3_ssRNAdelMn_chB_B_s2 |
| 2jlw_Deng4_NS3_ssRNA2_chA_s2 |
| 2jlw_Deng4_NS3_ssRNA2_chB_s2 |
| 2jlx_DV4_NS3hlcs_RNAdelMnChA_s2 |
| 2jlx_DV4_NS3hlcs_RNAdelMnChB_s2 |
| 2jlx_DV4_NS3hlcs_ssRNA_chA_s2 |
| 2jlx_DV4_NS3hlcs_ssRNA_chB_s2 |
| 2jly_DV4_NS3hlcs_ssRNA_chA_A_s2 |
| 2jly_DV4_NS3hlcs_ssRNA_chA_B_s2 |
| 2jly_DV4_NS3hlcs_ssRNAdelMn_chA_A_s2 |
| 2jly_DV4_NS3hlcs_ssRNAdelMn_chA_B_s2 |
| 2jly_DV4_NS3hlcs_ssRNAdelMn_chB_s2 |
| 2jlz_DV4_NS3hlcs_ssRNA_chA_s2 |
| 2jlz_DV4_NS3_ssRNAdelMn_chA_s2 |
| 2whx_DV_NS3hlcsDelMn_PRwasBnd_s2 |
| 2whx_DV_NS3hlcs_PRwasPepBnd_s2 |
| 1yks_YFV_NS3hlcs_s2 |
| 2z83_JEV_NS3hlcs_s2 |
| 1a1v_HCV_NS3hlcs_ssDNA_s2 |
| 4wxp_HCV_NS3helcs_A_s2 |
| 3kqh_HCV_NS3hlcs_ssDNA_chA_s2 |
| 3kqh_HCV_NS3hlcs_ssDNA_chB_s2 |
| 3kqk_HCV_NS3hlcs_chA_s2 |
| 3kqk_HCV_NS3hlcs_chB_s2 |
| 3kql_HCV_NS3hlcs_chnA_s2 |
| 3kql_HCV_NS3hlcs_chnB_s2 |
| 3kql_HCV_NS3hlcsDelMg_chA_s2 |
| 3kql_HCV_NS3hlcsDelMgChB_s2 |
| 3kqn_HCV_NS3hlcs_ssDNA_DelMn_s2 |
| 3kqn_HCV_NS3hlcs_ssDNA_s2 |
| 3o8b_HCV_NS3hlcs_PR_chA_s2 |
| 3o8b_HCV_NS3hlcs_PR_chB_s2 |
| 3o8c_HCV_NS3_Hlcs_PR_chB_s2 |
| 3o8c_HCV_NS3_ssRNA_wPR_chA_s2 |
| 1nkt_Mtb_SecA_chnA_s2 |
| 1nkt_Mtb_SecA_chnB_s2 |
| 1nkt_Mtb_SecA_DelMg_chnA_s2 |
| 1nkt_Mtb_SecA_DelMg_chnB_s2 |
| 4uaq_Mtb_SecA2_s2 |
| 4es6_PAeru_HemD_A_s2 |
| 4es6_PAeru_HemD_B_s2 |
| 4nl4_Klebp_PriA_A_s2 |
| 4nl4_Klebp_PriA_B_s2 |
| 3mla_Anthrax_BaNadDApochB_s2 |
| 3mla_Anthrax_BaNadD_chA_A_s2 |
| 3mla_Anthrax_BaNadD_chA_B_s2 |
| Batch Range | LibraryNickname |
| 90535 – 90543 | Drugs_and_Leads_FromPrestwick_MicroSourceSpectrum_FDA_and_NIHclinicalCollection |
| 90544 – 92658 | Enamine_Full_Library |
| 92659 – 94017 | Vitas-M Labs_Full_Library |
| 94018 – 94935 | ChemBridge_Full_Library |
| 94936 – 95394 | Asinex_Full_Library |
| 95395 – 95682 | NCI_Full_Library |
| 95683 – 95988 | Otava_Full_Library (note: now has 10,000 ligands/batch) |
Targets employed to virtual screening:
| x5gjb_ZIKV_NS3helicase_w_ssRNA.pdbqt |
| x5gjc_ZIKV_NS3helicase_wMn.pdbqt |
| x5jrz_NS3_ZIKV_FrPolynesia_A.pdbqt |
| x5jrz_NS3_ZIKV_FrPolynesia_B.pdbqt |
| x5gjc_ZIKV_NS3helicase_DelMn_s2.pdbqt |
| x5gjc_ZIKV_NS3helicase_wMn_s2.pdbqt |
| x5jrz_NS3_ZIKV_FrPolynesia_A_s2.pdbqt |
| x5jrz_NS3_ZIKV_FrPolynesia_B_s2.pdbqt |
| x5gjb_ZIKV_NS3helicase_w_ssRNA_s2.pdbqt |
Experiment 3A = Virtual Screening against the NS1 class (first to fifth binding site)
Glycoprotein NS1, present in all flaviviruses, appears to be essential for virus viability. This protein contains two conserved N-glycosylation sites and 12 invariant cysteine residues. The dockings were performed for five binding sites.
| Batch Range | LibraryNickname |
| 95989 – 96046 | Drugs_and_Leads_FromPrestwick_MicroSourceSpectrum_FDA_and_NIHclinicalCollection |
| 96047 – 109676 | Enamine_Full_Library |
| 109677 – 118434 | Vitas-M Labs_Full_Library |
| 118435 – 124350 | ChemBridge_Full_Library |
| 124351 – 127308 | Asinex_Full_Library |
| 127309 – 129164 | NCI_Full_Library |
| 129165 – 131136 | Otava_Full_Library (note: now has 10,000 ligands/batch) |
Targets employed to virtual screening:
| x5k6k_NS1_ZIKV_cA_plr1.pdbqt |
| x5k6k_NS1_ZIKV_cA_plr2.pdbqt |
| x5k6k_NS1_ZIKV_cA_NclcA5.pdbqt |
| x5k6k_NS1_ZIKV_cA_phL4.pdbqt |
| x5k6k_NS1_ZIKV_cA_phB3.pdbqt |
| x5k6k_NS1_ZIKV_cB_plr1.pdbqt |
| x5k6k_NS1_ZIKV_cB_plr2.pdbqt |
| x5k6k_NS1_ZIKV_cB_NclcA5.pdbqt |
| x5k6k_NS1_ZIKV_cB_phL4.pdbqt |
| x5k6k_NS1_ZIKV_cB_phB3.pdbqt |
| x5iy3_NS1_ZIKV_plr1.pdbqt |
| x5iy3_NS1_ZIKV_plr2.pdbqt |
| x5iy3_NS1_ZIKV_phL4.pdbqt |
| x5iy3_NS1_ZIKV_phB3.pdbqt |
| x4o6b_NS1_Dengue_plr1.pdbqt |
| x4o6d_NS1_WNV_Holo_plr1.pdbqt |
| x4o6d_NS1_WNV_wWATs_plr1.pdbqt |
| x4oie_NS1_WNV_Holo_plr1.pdbqt |
| x4oie_NS1_WNV_wWATs_plr1.pdbqt |
| x4oig_NS1_Dengue_AB_plr1.pdbqt |
| x4oig_NS1_Dengue_DE_plr1.pdbqt |
| x4oii_NS1_WNV_plr1.pdbqt |
| x4tpl_NS1_WNV_plr1.pdbqt |
| x4o6b_NS1_Dengue_plr2.pdbqt |
| x4o6d_NS1_WNV_Holo_plr2.pdbqt |
| x4o6d_NS1_WNV_wWATs_plr2.pdbqt |
| x4oie_NS1_WNV_Holo_plr2.pdbqt |
| x4oie_NS1_WNV_wWATs_plr2.pdbqt |
| x4oig_NS1_Dengue_AB_plr2.pdbqt |
| x4oig_NS1_Dengue_DE_plr2.pdbqt |
| x4oii_NS1_WNV_plr2.pdbqt |
| x4tpl_NS1_WNV_plr2.pdbqt |
| x4o6b_NS1_Dengue_NclcA5.pdbqt |
| x4o6d_NS1_WNV_Holo_NclcA5.pdbqt |
| x4o6d_NS1_WNV_wWATs_NclcA5.pdbqt |
| x4tpl_NS1_WNV_NclcA5.pdbqt |
| x4o6b_NS1_Dengue_phL4.pdbqt |
| x4o6d_NS1_WNV_Holo_phL4.pdbqt |
| x4o6d_NS1_WNV_wWATs_phL4.pdbqt |
| x4oie_NS1_WNV_Holo_phL4.pdbqt |
| x4oie_NS1_WNV_wWATs_phL4.pdbqt |
| x4oig_NS1_Dengue_AB_phL4.pdbqt |
| x4oig_NS1_Dengue_DE_phL4.pdbqt |
| x4oii_NS1_WNV_phL4.pdbqt |
| x4tpl_NS1_WNV_phL4.pdbqt |
| x4o6b_NS1_Dengue_phB3.pdbqt |
| x4o6d_NS1_WNV_Holo_phB3.pdbqt |
| x4o6d_NS1_WNV_wWATs_phB3.pdbqt |
| x4oie_NS1_WNV_Holo_phB3.pdbqt |
| x4oie_NS1_WNV_wWATs_phB3.pdbqt |
| x4oig_NS1_Dengue_AB_phB3.pdbqt |
| x4oig_NS1_Dengue_DE_phB3.pdbqt |
| x4oii_NS1_WNV_phB3.pdbqt |
| x4tpl_NS1_WNV_phB3.pdbqt |
| x3lv2_DTBS_BioA_Mtb_A.pdbqt |
| x3lv2_DTBS_BioA_Mtb_A_s2.pdbqt |
| x3lv2_DTBS_BioA_Mtb_B.pdbqt |
| x3lv2_DTBS_BioA_Mtb_B_s2.pdbqt |
Experiment 4A = Virtual Screening against the NS2B-NS3 protease class
NS2B-NS3 protease is responsible for all cytoplasmic cleavages including at junctions between NS2A/NS2B, NS2B/NS3, NS3/NS4A and NS4B/NS5 proteins and within the capsid, NS2A and NS4A proteins. Similar to NS3-NS4A protease from hepatitis C virus, the flavivirus NS2B-NS3 protease is essential for the virus replicative cycle, and thus constitutes an ideal target for antiviral drug development. NS2B-NS3 protease also suppresses the immune response by cleaving stimulator of interferon genes (STING) in DENV, triggers apoptosis via activating caspases in WNV, and induces neurotropic pathogenesis by inhibiting activator protein 1 (AP-1) in JEV.
| Batch Range | LibraryNickname |
| 166891 – 166960 | Drugs_and_Leads_FromPrestwick_MicroSourceSpectrum_FDA_and_NIHclinicalCollection |
| 166961 – 183410 | Enamine_Full_Library |
| 183411 – 193980 | Vitas-M Labs_Full_Library |
| 193981 – 201120 | ChemBridge_Full_Library |
| 201121 – 204690 | Asinex_Full_Library |
| 204691 – 206930 | NCI_Full_Library |
| 206931 – 209310 | Otava_Full_Library (note: now has 10,000 ligands/batch) |
Targets employed to virtual screening:
| x5gj4_NS2BNS3pr_ZIKV_A_AB.pdbqt | ||
| x5h4i_ZIKV_NS2bNS3pr_HadFrag.pdbqt | ||
| x5lc0_ZIKV_NS2BNS3pr_Bo_chnA.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_A_CD.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_A_EF.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_A_GH.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_A_AB.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_A_CD.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_A_EF.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_A_GH.pdbqt | ||
| x5t1v_NS2B_NS3pr_ZIKV_apo_chnA.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_A_AB_DelNS2b.pdbqt | ||
| x5h4i_ZIKV_NS2bNS3pr_DelNS2b.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_A_CD_DelNS2b.pdbqt | ||
| x5t1v_NS2bNS3_ZIKV_apoChnA_DelNS2b.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_A_EF_DelNS2b.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_A_GH_DelNS2b.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_B_AB.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_B_CD.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_B_EF.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_B_GH.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_B_AB.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_B_CD.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_B_EF.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_B_GH.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_B_AB_delNS2b.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_B_CD_delNS2b.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_B_EF_delNS2b.pdbqt | ||
| x5gj4_NS2BNS3pr_ZIKV_B_GH_delNS2b.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_A_AB_DelNS2m.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_A_CD_DelNS2m.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_A_EF_DelNS2m.pdbqt | ||
| x5gpi_ZIKV_NS2bNS3pr_A_GH_DelNS2m.pdbqt | ||
| x5lc0_ZIKV_NS2BNS3pr_Bo_chnB.pdbqt | ||
| x5lc0_ZIKV_NS2BNS3pr_Dimer.pdbqt | ||
| x5t1v_NS2B_NS3pr_ZIKV_apo_chnB.pdbqt | ||
| x5t1v_NS2bNS3_ZIKV_apoChnB_DelNS2b.pdbqt | ||
| x2fom_Deng_NS2bNS3pr_A.pdbqt | ||
| x2fom_Deng_NS2bNS3pr_dimer_B.pdbqt | ||
| x2fp7_Deng_NS2bNS3pr_wPepInhib.pdbqt | ||
| x2ijo_WNV_NS2bNS3pr_wBPTI.pdbqt | ||
| x2m9p_NMR10_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9p_NMR1_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9p_NMR2_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9p_NMR3_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9p_NMR4_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9p_NMR5_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9p_NMR6_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9p_NMR7_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9p_NMR8_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9p_NMR9_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_10NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_1NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_2NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_3NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_4NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_5NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_6NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_7NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_8NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2m9q_9NMR_DengNS3pr_wPepAnalog.pdbqt | ||
| x2whx_Dengue_NS3NS2Bpr_DelNS2b.pdbqt | ||
| x2whx_Dengue_NS3NS2Bprotease.pdbqt | ||
| x3o8b_HCV_NS2BNS3protease_chA.pdbqt | ||
| x3o8b_HCV_NS2BNS3protease_chB.pdbqt | ||
| x3o8c_HCV_NS2BNS3protease_chA.pdbqt | ||
| x3o8c_HCV_NS2BNS3protease_chB.pdbqt | ||
| x1qy6_Saur_V8pr.pdbqt | ||
| x1wcz_Saur_V8pr.pdbqt | ||
| x2as9_Saur_SpI_pr_1.pdbqt |
Experiment 4B = Virtual Screening against the NS2B-NS3 protease class
| Batch Range | LibraryNickname |
| 209311 – 209401 | Drugs_and_Leads_FromPrestwick_MicroSourceSpectrum_FDA_and_NIHclinicalCollection |
| 209402 – 230786 | Enamine_Full_Library |
| 230787 – 244527 | Vitas-M Labs_Full_Library |
| 244528 – 253809 | ChemBridge_Full_Library |
| 253810 – 258450 | Asinex_Full_Library |
| 258451 – 261362 | NCI_Full_Library |
| 261363 – 264456 | Otava_Full_Library (note: now has 10,000 ligands/batch) |
Targets employed to virtual screening:
| x3u1j_Deng_NS2bNS3pr_Aprtnn_A.pdbqt |
| x4m9t_DengNS2bNS3pr_CtoA_PAllos_A.pdbqt |
| x4m9t_DengNS2bNS3pr_CtoA_PAllos_B.pdbqt |
| x4m9f_DengNS2bNS3pr_CmodtoA_A.pdbqt |
| x4m9f_DengNS2bNS3pr_CmodtoA_B.pdbqt |
| x4m9k_DengNS2bNS3pr_pH5.5_A.pdbqt |
| x4m9k_DengNS2bNS3pr_pH5.5_B.pdbqt |
| x4m9k_DengNS2bNS3pr_pH5.5_C.pdbqt |
| x4m9m_DengNS2bNS3pr_pH8.5_A.pdbqt |
| x4m9m_DengNS2bNS3pr_pH8.5_B.pdbqt |
| x4m9m_DengNS2bNS3pr_pH8.5_C.pdbqt |
| x2yol_WNV_NS2bNS3pr_hadInhib_A.pdbqt |
| x2yol_WNV_NS2bNS3pr_hadInhib_B.pdbqt |
| x3e90_WNV_NS2bNS3pr_Pep_AB_A.pdbqt |
| x3e90_WNV_NS2bNS3pr_Pep_AB_B.pdbqt |
| x3e90_WNV_NS2bNS3pr_Pep_CD_A.pdbqt |
| x3e90_WNV_NS2bNS3pr_Pep_CD_B.pdbqt |
| x2vbc_Deng_fullNS3prNhel.pdbqt |
| x2wv9_MVEV_NS3pr_chB.pdbqt |
| x4r8t_JEV_NS2bPR.pdbqt |
| x4wf8_HCV_NS3pr_Asnprvr_A.pdbqt |
| x4wf8_HCV_NS3pr_Asnprvr_B.pdbqt |
| x4wh8_1_HCV_NS3pr_CycAsnprvr.pdbqt |
| x4wh8_2_HCV_NS3pr_CycAsnprvr.pdbqt |
| x5epn_HCV_NS3pr_MK5172cyc.pdbqt |
| x5eqq_HCV_NS3pr_MK5172linear_A.pdbqt |
| x5eqq_HCV_NS3pr_MK5172linear_B.pdbqt |
| x4nwk_HCV_NS3pr_BMS339_A.pdbqt |
| x4nwk_HCV_NS3pr_BMS339_B.pdbqt |
| x4nwl_HCV_NS3pr_BMS032_A.pdbqt |
| x4nwl_HCV_NS3pr_BMS032_B.pdbqt |
| x3lon_HCV_NS3pr_Nrlprvr_A.pdbqt |
| x3lox_HCV_NS3pr_Boceprevir.pdbqt |
| x3p8n_HCV_NS3pr_BIinhib_chA.pdbqt |
| x3p8n_HCV_NS3pr_BIinhib_chB.pdbqt |
| x3su3_HCV_NS3pr_Vaniprevir_A.pdbqt |
| x3su3_HCV_NS3pr_Vaniprevir_B.pdbqt |
| x3su3_HCV_NS3pr_Vaniprevir_C.pdbqt |
| x3sv6_HCV_NS3pr_Telaprevir_A.pdbqt |
| x3sv6_HCV_NS3pr_Telaprevir_B.pdbqt |
| x3sv6_HCV_NS3pr_Telaprevir_C.pdbqt |
| x3sv6_HCV_NS3pr_Telaprevir_D.pdbqt |
| x3sv8_HCV_NS3prD168A_Tlprvr_A.pdbqt |
| x3sv9_HCV_NS3prA156T_Tlprvr_A.pdbqt |
| x3sv9_HCV_NS3prA156T_Tlprvr_B.pdbqt |
| x3sv9_HCV_NS3prA156T_Tlprvr_C.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_A_A.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_A_B.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_B_A.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_C_A.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_C_B.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_D_A.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_D_B.pdbqt |
| x3lon_HCV_NS3pr_apo_chC.pdbqt |
| x4a1v_HCV_NS3pr_OptPep_A.pdbqt |
| x4a1v_HCV_NS3pr_OptPep_B.pdbqt |
| x4a1x_HCV_NS3pr_PepCP546A_A_A.pdbqt |
| x4a1x_HCV_NS3pr_PepCP546A_A_B.pdbqt |
| x4a1x_HCV_NS3pr_PepCP546A_B_A.pdbqt |
| x4a1x_HCV_NS3pr_PepCP546A_B_B.pdbqt |
| x4i31_HCV_NS3pr_Macrocyc_A.pdbqt |
| x4i31_HCV_NS3pr_Macrocyc_B.pdbqt |
| x4jmy_HCV_NS3pr_DDIVPC_A.pdbqt |
| x4jmy_HCV_NS3pr_DDIVPC_B.pdbqt |
| x4ktc_HCV_NS3pr_Macrocyc1nm_A.pdbqt |
| x4ktc_HCV_NS3pr_Macrocyc1nm_C.pdbqt |
| x4u01_1_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_2_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_3_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_4_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_5_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_6_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_7_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_8_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_9_HCV_NS3pr_Macrocyc70.pdbqt |
| x3ufa_Saur_SplApr_PInhib_A.pdbqt |
| x3ufa_Saur_SplApr_PInhib_B.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chA_A.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chA_B.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chB_A.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chB_B.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chC_A.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chC_B.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chD_A.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chD_B.pdbqt |
| x2w7s_1_A_Saur_SplA_pr.pdbqt |
| x2w7s_1_B_Saur_SplA_pr.pdbqt |
| x2w7s_3_A_Saur_SplA_pr.pdbqt |
| x2w7s_3_B_Saur_SplA_pr.pdbqt |
| x2w7s_4_Saur_SplA_pr.pdbqt |
| x2o8l_Saur_V8pr.pdbqt |
Experiment 4C = Virtual Screening against the NS2B-NS3 protease class
| Batch Range | Library Nickname |
| 264457 – 292511 | BigLib (ZINC15 Library) |
Targets employed to virtual screening:
| x3u1j_Deng_NS2bNS3pr_Aprtnn_A.pdbqt |
| x4m9t_DengNS2bNS3pr_CtoA_PAllos_A.pdbqt |
| x4m9t_DengNS2bNS3pr_CtoA_PAllos_B.pdbqt |
| x4m9f_DengNS2bNS3pr_CmodtoA_A.pdbqt |
| x4m9f_DengNS2bNS3pr_CmodtoA_B.pdbqt |
| x4m9k_DengNS2bNS3pr_pH5.5_A.pdbqt |
| x4m9k_DengNS2bNS3pr_pH5.5_B.pdbqt |
| x4m9k_DengNS2bNS3pr_pH5.5_C.pdbqt |
| x4m9m_DengNS2bNS3pr_pH8.5_A.pdbqt |
| x4m9m_DengNS2bNS3pr_pH8.5_B.pdbqt |
| x4m9m_DengNS2bNS3pr_pH8.5_C.pdbqt |
| x2yol_WNV_NS2bNS3pr_hadInhib_A.pdbqt |
| x2yol_WNV_NS2bNS3pr_hadInhib_B.pdbqt |
| x3e90_WNV_NS2bNS3pr_Pep_AB_A.pdbqt |
| x3e90_WNV_NS2bNS3pr_Pep_AB_B.pdbqt |
| x3e90_WNV_NS2bNS3pr_Pep_CD_A.pdbqt |
| x3e90_WNV_NS2bNS3pr_Pep_CD_B.pdbqt |
| x2vbc_Deng_fullNS3prNhel.pdbqt |
| x2wv9_MVEV_NS3pr_chB.pdbqt |
| x4r8t_JEV_NS2bPR.pdbqt |
| x4wf8_HCV_NS3pr_Asnprvr_A.pdbqt |
| x4wf8_HCV_NS3pr_Asnprvr_B.pdbqt |
| x4wh8_1_HCV_NS3pr_CycAsnprvr.pdbqt |
| x4wh8_2_HCV_NS3pr_CycAsnprvr.pdbqt |
| x5epn_HCV_NS3pr_MK5172cyc.pdbqt |
| x5eqq_HCV_NS3pr_MK5172linear_A.pdbqt |
| x5eqq_HCV_NS3pr_MK5172linear_B.pdbqt |
| x4nwk_HCV_NS3pr_BMS339_A.pdbqt |
| x4nwk_HCV_NS3pr_BMS339_B.pdbqt |
| x4nwl_HCV_NS3pr_BMS032_A.pdbqt |
| x4nwl_HCV_NS3pr_BMS032_B.pdbqt |
| x3lon_HCV_NS3pr_Nrlprvr_A.pdbqt |
| x3lox_HCV_NS3pr_Boceprevir.pdbqt |
| x3p8n_HCV_NS3pr_BIinhib_chA.pdbqt |
| x3p8n_HCV_NS3pr_BIinhib_chB.pdbqt |
| x3su3_HCV_NS3pr_Vaniprevir_A.pdbqt |
| x3su3_HCV_NS3pr_Vaniprevir_B.pdbqt |
| x3su3_HCV_NS3pr_Vaniprevir_C.pdbqt |
| x3sv6_HCV_NS3pr_Telaprevir_A.pdbqt |
| x3sv6_HCV_NS3pr_Telaprevir_B.pdbqt |
| x3sv6_HCV_NS3pr_Telaprevir_C.pdbqt |
| x3sv6_HCV_NS3pr_Telaprevir_D.pdbqt |
| x3sv8_HCV_NS3prD168A_Tlprvr_A.pdbqt |
| x3sv9_HCV_NS3prA156T_Tlprvr_A.pdbqt |
| x3sv9_HCV_NS3prA156T_Tlprvr_B.pdbqt |
| x3sv9_HCV_NS3prA156T_Tlprvr_C.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_A_A.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_A_B.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_B_A.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_C_A.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_C_B.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_D_A.pdbqt |
| x3kee_HCV_NS3NS4aPR_TMC435_D_B.pdbqt |
| x3lon_HCV_NS3pr_apo_chC.pdbqt |
| x4a1v_HCV_NS3pr_OptPep_A.pdbqt |
| x4a1v_HCV_NS3pr_OptPep_B.pdbqt |
| x4a1x_HCV_NS3pr_PepCP546A_A_A.pdbqt |
| x4a1x_HCV_NS3pr_PepCP546A_A_B.pdbqt |
| x4a1x_HCV_NS3pr_PepCP546A_B_A.pdbqt |
| x4a1x_HCV_NS3pr_PepCP546A_B_B.pdbqt |
| x4i31_HCV_NS3pr_Macrocyc_A.pdbqt |
| x4i31_HCV_NS3pr_Macrocyc_B.pdbqt |
| x4jmy_HCV_NS3pr_DDIVPC_A.pdbqt |
| x4jmy_HCV_NS3pr_DDIVPC_B.pdbqt |
| x4ktc_HCV_NS3pr_Macrocyc1nm_A.pdbqt |
| x4ktc_HCV_NS3pr_Macrocyc1nm_C.pdbqt |
| x4u01_1_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_2_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_3_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_4_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_5_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_6_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_7_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_8_HCV_NS3pr_Macrocyc70.pdbqt |
| x4u01_9_HCV_NS3pr_Macrocyc70.pdbqt |
| x3ufa_Saur_SplApr_PInhib_A.pdbqt |
| x3ufa_Saur_SplApr_PInhib_B.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chA_A.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chA_B.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chB_A.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chB_B.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chC_A.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chC_B.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chD_A.pdbqt |
| x4mvn_Saur_SplApr_Inhib_chD_B.pdbqt |
| x2w7s_1_A_Saur_SplA_pr.pdbqt |
| x2w7s_1_B_Saur_SplA_pr.pdbqt |
| x2w7s_3_A_Saur_SplA_pr.pdbqt |
| x2w7s_3_B_Saur_SplA_pr.pdbqt |
| x2w7s_4_Saur_SplA_pr.pdbqt |
| x2o8l_Saur_V8pr.pdbqt |
Experiment 2E = Virtual Screening against the NS3 helicase class (new structure)
| Batch Range | LibraryNickname |
| 292512 – 319661 | BigLib |
Targets employed to virtual screening:
|
x5gjb_ZIKV_NS3h_w_RNA_1b
|
|
x5gjb_ZIKV_NS3h_w_RNA_MDavg_1b
|
|
x5mfx_ZIKV_NS3h_w_RNA_A_1b
|
|
x5mfx_ZIKV_NS3h_w_RNA_B_1b
|
|
x5gjb_ZIKV_NS3h_MD102closed_s2b
|
|
x5jmt_ZIKV_NS3h_MD23Open_s2b
|
|
x5k8l_ZIKV_NS3_AlsTarget_A_s2b
|
|
x5k8l_ZIKV_NS3_AlsTarget_A_w9WATs_s2b
|
|
x5gjb_ZIKV_NS3h_w_RNA_s2b
|
|
x5gjc_ZIKV_NS3h_DelMn_s2b
|
|
x5gjc_ZIKV_NS3h_wMn_s2b
|
|
x5jmt_ZIKV_NS3h_A_s2b
|
|
x5k8i_ZIKV_NS3h_DelMn_A_s2b
|
|
x5k8i_ZIKV_NS3h_wMg_A_s2b
|
|
x5k8t_ZIKV_NS3_A_DelMgs_s2b
|
|
x5k8t_ZIKV_NS3_A_w1Mg_s2b
|
|
x5k8u_ZIKV_NS3_A_DelMgs_s2b
|
|
x5k8u_ZIKV_NS3_A_wMg_s2b
|
|
x5k8u_ZIKV_NS3_A_wMg_w4WATs_s2b
|
|
x5mfx_ZIKV_NS3h_w_RNA_A_s2b
|
|
x5y4z_ZIKV_NS3hel_hadANP_s2b
|
|
x2jlx_DV4_NS3hlcs_ssRNA_chA_s2b
|
|
x2jly_DV4_NS3hlcs_ssRNA_chA_A_s2b
|
|
x3kql_HCV_NS3hlcs_chnB_s2b
|
|
x3kqn_HCV_NS3hlcs_ssDNA_s2b
|
|
x1nkt_Mtb_SecA_chnA
|
|
x4es6_PAeru_HemD_A
|
|
x4nl4_Klebp_PriA_A
|
|
x1nkt_Mtb_SecA_chnA_s2b
|
|
x4nl4_Klebp_PriA_A_s2b
|
Experiment 1C = Virtual Screening against the NS5 class (methyltransferase and full length)
| Batch Range | LibraryNickname |
| 292512 – 319661 | BigLib |
Targets employed to virtual screening:
|
x5gjb_ZIKV_NS3h_w_RNA_1b
|
|
x5gjb_ZIKV_NS3h_w_RNA_MDavg_1b
|
|
x5mfx_ZIKV_NS3h_w_RNA_A_1b
|
|
x5mfx_ZIKV_NS3h_w_RNA_B_1b
|
|
x5gjb_ZIKV_NS3h_MD102closed_s2b
|
|
x5jmt_ZIKV_NS3h_MD23Open_s2b
|
|
x5k8l_ZIKV_NS3_AlsTarget_A_s2b
|
|
x5k8l_ZIKV_NS3_AlsTarget_A_w9WATs_s2b
|
|
x5gjb_ZIKV_NS3h_w_RNA_s2b
|
|
x5gjc_ZIKV_NS3h_DelMn_s2b
|
|
x5gjc_ZIKV_NS3h_wMn_s2b
|
|
x5jmt_ZIKV_NS3h_A_s2b
|
|
x5k8i_ZIKV_NS3h_DelMn_A_s2b
|
|
x5k8i_ZIKV_NS3h_wMg_A_s2b
|
|
x5k8t_ZIKV_NS3_A_DelMgs_s2b
|
|
x5k8t_ZIKV_NS3_A_w1Mg_s2b
|
|
x5k8u_ZIKV_NS3_A_DelMgs_s2b
|
|
x5k8u_ZIKV_NS3_A_wMg_s2b
|
|
x5k8u_ZIKV_NS3_A_wMg_w4WATs_s2b
|
|
x5mfx_ZIKV_NS3h_w_RNA_A_s2b
|
|
x5y4z_ZIKV_NS3hel_hadANP_s2b
|
|
x2jlx_DV4_NS3hlcs_ssRNA_chA_s2b
|
|
x2jly_DV4_NS3hlcs_ssRNA_chA_A_s2b
|
|
x3kql_HCV_NS3hlcs_chnB_s2b
|
|
x3kqn_HCV_NS3hlcs_ssDNA_s2b
|
|
x1nkt_Mtb_SecA_chnA
|
|
x4es6_PAeru_HemD_A
|
|
x4nl4_Klebp_PriA_A
|
|
x1nkt_Mtb_SecA_chnA_s2b
|
|
x4nl4_Klebp_PriA_A_s2b
|
Experiment 5A = Virtual Screening against the Capsid protein (binding pocket 1)
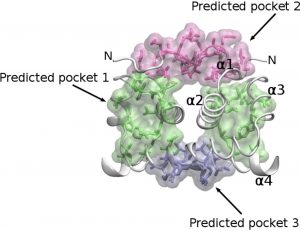
| Batch Range | LibraryNickname |
| 338667 -364911 | BigLib (ZINC15 library) |
Targets employed to virtual screening:
| x1r6r_DENV2_capsid_NMR_10 |
| x1r6r_DENV2_capsid_NMR_4 |
| x1r6r_DENV2_capsid_NMR_11 |
| x1r6r_DENV2_capsid_NMR_5 |
| x1r6r_DENV2_capsid_NMR_12 |
| x1r6r_DENV2_capsid_NMR_6 |
| x1r6r_DENV2_capsid_NMR_13 |
| x1r6r_DENV2_capsid_NMR_7 |
| x1r6r_DENV2_capsid_NMR_14 |
| x1r6r_DENV2_capsid_NMR_8 |
| x1r6r_DENV2_capsid_NMR_15 |
| x1r6r_DENV2_capsid_NMR_9 |
| x1r6r_DENV2_capsid_NMR_16 |
| x1sfk_WNV_Capsid_3_2A_chAB |
| x1r6r_DENV2_capsid_NMR_17 |
| x1sfk_WNV_Capsid_3_2A_chCD |
| x1r6r_DENV2_capsid_NMR_18 |
| x1sfk_WNV_Capsid_3_2A_chEF |
| x1r6r_DENV2_capsid_NMR_19 |
| x1sfk_WNV_Capsid_3_2A_chGH |
| x1r6r_DENV2_capsid_NMR_1 |
| x5ygh_ZIKV_Capsid_1_88A |
| x1r6r_DENV2_capsid_NMR_20 |
| x5z0r_ZIKV_Capsid |
| x1r6r_DENV2_capsid_NMR_21 |
| x5z0v_ZIKV_Capsid1 |
| x1r6r_DENV2_capsid_NMR_2 |
| x5z0v_ZIKV_Capsid2 |
| x1r6r_DENV2_capsid_NMR_3 |
Experiment 5B = Virtual Screening against the Capsid protein (binding pocket 2)
| Batch Range | LibraryNickname |
| 364912 -391156 | BigLib (ZINC15 library) |
Targets employed to virtual screening:
| x1r6r_DENV2_capsid_NMR_10_s2 |
| x1r6r_DENV2_capsid_NMR_4_s2 |
| x1r6r_DENV2_capsid_NMR_11_s2 |
| x1r6r_DENV2_capsid_NMR_5_s2 |
| x1r6r_DENV2_capsid_NMR_12_s2 |
| x1r6r_DENV2_capsid_NMR_6_s2 |
| x1r6r_DENV2_capsid_NMR_13_s2 |
| x1r6r_DENV2_capsid_NMR_7_s2 |
| x1r6r_DENV2_capsid_NMR_14_s2 |
| x1r6r_DENV2_capsid_NMR_8_s2 |
| x1r6r_DENV2_capsid_NMR_15_s2 |
| x1r6r_DENV2_capsid_NMR_9_s2 |
| x1r6r_DENV2_capsid_NMR_16_s2 |
| x1sfk_WNV_capsid_3_2A_chAB_s2 |
| x1r6r_DENV2_capsid_NMR_17_s2 |
| x1sfk_WNV_capsid_3_2A_chCD_s2 |
| x1r6r_DENV2_capsid_NMR_18_s2 |
| x1sfk_WNV_capsid_3_2A_chEF_s2 |
| x1r6r_DENV2_capsid_NMR_19_s2 |
| x1sfk_WNV_capsid_3_2A_chGH_s2 |
| x1r6r_DENV2_capsid_NMR_1_s2 |
| x5ygh_ZIKV_capsid_1_88A_s2 |
| x1r6r_DENV2_capsid_NMR_20_s2 |
| x5z0r_ZIKV_capsid_s2 |
| x1r6r_DENV2_capsid_NMR_21_s2 |
| x5z0v_ZIKV_capsid1_s2 |
| x1r6r_DENV2_capsid_NMR_2_s2 |
| x5z0v_ZIKV_capsid2_s2 |
| x1r6r_DENV2_capsid_NMR_3_s2 |
Experiment 6A = Virtual Screening against the Envelope protein
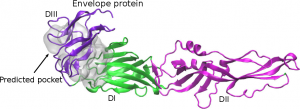
| Batch Range | LibraryNickname |
| 391157-404731 | BigLib (ZINC15 library) |
Targets employed to virtual screening:
| x3j2p_DENV2_E |
| x5kve_ZIKV_E_DIII-antibody |
| x3uaj_DENV4_E-antibody |
| x5kvf_ZIKV_E_DIII-antibody |
| x4uta_DENV2_E-antibody |
| x5kvg_ZIKV_E_DIII-antibody |
| x5gzn_ZIKV_E-antibody |
| x5lbs_ZIKV_E-antibody |
| x5gzo_ZIKV_E-antibody |
| x5lbv_ZIKV_E-antibody |
| x5jhl_ZIKV_E-antibody |
| x5vig_ZIKV_E_DIII_antibody |
| x5jhm_ZIKV_E |
| x6dfi_ZIKV_E_DIII-antibody |
| x5kvd_ZIKV_E_DIII-antibody |
Experiment 3B = Virtual Screening against the NS1 protein (crystal and protein conformations from MD simulation) – site 1
| Batch Range | LibraryNickname |
| 404732-412876 | BigLib (ZINC15 library) |
Targets employed to virtual screening:
| x5k6k_ZIKV_NS1_MD_model_1_s1 |
| x5k6k_ZIKV_NS1_MD_model_6_s1 |
| x5k6k_ZIKV_NS1_MD_model_2_s1 |
| x5k6k_ZIKV_NS1_MD_model_7_s1 |
| x5k6k_ZIKV_NS1_MD_model_3_s1 |
| x5k6k_ZIKV_NS1_MD_model_8_s1 |
| x5k6k_ZIKV_NS1_MD_model_4_s1 |
| x5k6k_ZIKV_NS1_s1 |
| x5k6k_ZIKV_NS1_MD_model_5_s1 |
Experiment 3C = Virtual Screening against the NS1 protein (crystal and protein conformations from MD simulation) – site 2
| Batch Range | LibraryNickname |
| 412877-421021 | BigLib (ZINC15 library) |
Targets employed to virtual screening:
| x5k6k_ZIKV_NS1_MD_model_1_s2 |
| x5k6k_ZIKV_NS1_MD_model_6_s2 |
| x5k6k_ZIKV_NS1_MD_model_2_s2 |
| x5k6k_ZIKV_NS1_MD_model_7_s2 |
| x5k6k_ZIKV_NS1_MD_model_3_s2 |
| x5k6k_ZIKV_NS1_MD_model_8_s2 |
| x5k6k_ZIKV_NS1_MD_model_4_s2 |
| x5k6k_ZIKV_NS1_s2 |
| x5k6k_ZIKV_NS1_MD_model_5_s2 |
Experiment 1D = Virtual Screening against the NS5 polymerase protein (NTP site)
| Batch Range | LibraryNickname |
| 421022-444551 | BigLib (ZINC15 library) |
Targets employed to virtual screening:
| x2j7u_DENV_NS5pol_s1 |
| x5hmz_DENV3_NS5_pol_s1 |
| x2j7w_DENV_NS5pol_s1 |
| x5hn0_DENV3_NS5pol_s1 |
| x3vws_DENV_NS5pol_s1 |
| x5i3p_DENV3_NS5pol_s1 |
| x4c11_DENV_NS5pol_s1 |
| x5i3q_DENV3_NS5pol_s1 |
| x4hhj_DENV3_NS5pol_s1 |
| x5iq6_DENV3_NS5pol_s1 |
| x4v0q_DENV_NS5_s1 |
| x5jjr_DENV3_NS5_s1 |
| x4v0r_DENV_NS5_s1 |
| x5jjs_DENV3_NS5_s1 |
| x5f3t_DENV3_NS5pol_s1 |
| x5k5m_DENV2_NS5pol_s1 |
| x5f3z_DENV3_NS5pol_s1 |
| x5tfr_ZIKV_NS5_s1 |
| x5f41_DENV3_NS5pol_s1 |
| x5u04_ZIKV_NS5pol_s1 |
| x5hmw_DENV3_NS5pol_s1 |
| x5wz3_ZIKV_NS5pol_s1 |
| x5hmx_DENV3_NS5pol_s1 |
| x6h80_DENV_NS5pol_s1 |
| x5hmy_DENV3_NS5pol_s1 |
| x6h9r_DENV3_NS5pol_s1 |
Experiment 1E = Virtual Screening against the NS5 polymerase protein (RNA site)
| Batch Range | LibraryNickname |
| 444552-452095 | BigLib (ZINC15 library) |
Targets employed to virtual screening:
| x5tfr_ZIKV_NS5_s3 |
| x5wz3_ZIKV_NS5pol_s3 |
| x5u04_ZIKV_NS5pol_s3 |
| x5jjs_DENV3_NS5_s3 |
| x4v0r_DENV_NS5_s3 |
| x2j7u_DENV_NS5pol_s3 |
| x5i3q_DENV3_NS5pol_s3 |
| x5hmz_DENV3_NS5_pol_s3 |
| x5jjr_DENV3_NS5_s3 |
Experiment 1F = Virtual Screening against the NS5 polymerase protein (Active binding site)
| Batch Range | LibraryNickname |
| 452096-459586 | BigLib (ZINC15 library) |
Targets employed to virtual screening:
| x5tfr_ZIKV_NS5_s2 |
| x5wz3_ZIKV_NS5pol_s2 |
| x5u04_ZIKV_NS5pol_s2 |
| x5jjs_DENV3_NS5_s2 |
| x4v0r_DENV_NS5_s2 |
| x2j7u_DENV_NS5pol_s2 |
| x5i3q_DENV3_NS5pol_s2 |
| x5hmz_DENV3_NS5_pol_s2 |
| x5jjr_DENV3_NS5_s2 |
Experiment 1H = Virtual Screening against the NS5 methyltransferase protein (GTP binding site)
| Batch Range | LibraryNickname |
| 459587-465016 | BigLib (ZINC15 library) |
Targets employed to virtual screening:
| x5kqr_NS5met_s3 | ||
| x5wxb_NS5met_s3 | ||
| x5mrk_NS5met_s3 | ||
| x5kqr_NS5met_MD_model1_s3 | ||
| x5kqr_NS5met_MD_model2_s3 | ||
| x5kqr_NS5met_MD_model3_s3 | ||





