Illustrating and homology modeling the proteins of the Zika virus
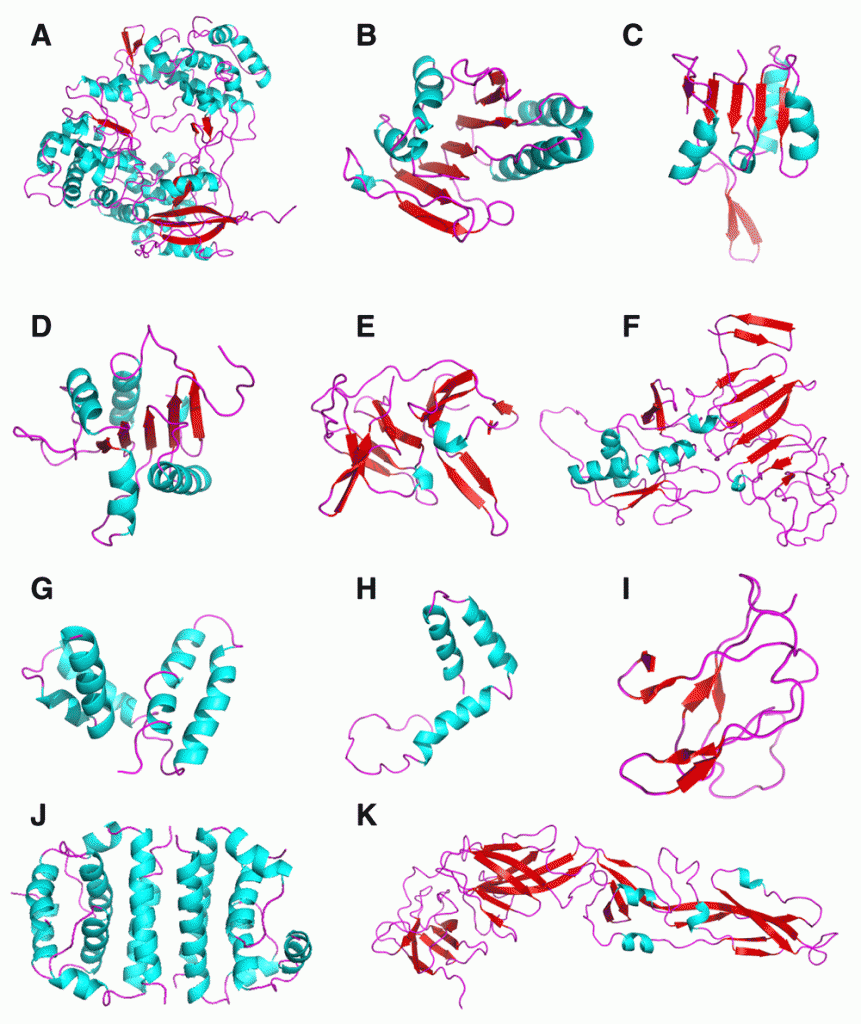
Several suggested steps have been proposed which could be taken to initiate ZIKV antiviral drug discovery using both high throughput screens as well as structure-based design based on homology models for the key proteins. We now describe preliminary homology models created for NS5, FtsJ, NS4B, NS4A, HELICc, DEXDc, peptidase S7, NS2B, NS2A, NS1, E stem, glycoprotein M, propeptide, capsid and glycoprotein E using SWISS-MODEL.
May 5, 2016
Illustrating and homology modeling the proteins of the Zika virus – F1000Research








Leave a Reply
Want to join the discussion?Feel free to contribute!